Ligandbook is a public repository for force field parameters with a special emphasis on small molecules and known ligands of proteins. It acts as a warehouse for parameter files that are supplied by the community. Anyone can browse and download parameters; deposition of files requires user registration.
Ligandbook's aims are to
In order to achieve these aims, Ligandbook provides a stable framework for any user to search and download deposited parameter sets. Users such as forcefield parameter authors can also upload their parameter sets (requires free registration). Parameter sets can be updated but even then older versions are still available and provide a transparent history of changes so that specific studies can be reproduced with the exact same parameters if needed.
Each parameter set is associated with a single chemical compound. Ligandbook can automatically recognize a compound on upload and link to entries in the PubChem and the Protein Databank.
Publications describing the parameterization can be added as references. By supplying a PubMed ID, citation data are automatically downloaded and linked to the PubMed entry. The citation data are always distributed together with the parameters themselves so that any user has all the information needed to read the original paper, ascertain appropriate use, and, importantly, to cite the original authors appropriately.
In order to further help users in validating parameters, Ligandbook also contains additional metadata that allows listing of experimental and computed observables together with the appropriate references. Together with the links to established databases of experimental data (PubChem, Protein Databank) and the references, a user is given as much information as possible to make an informed decision about the validity and applicability of a parameter set.
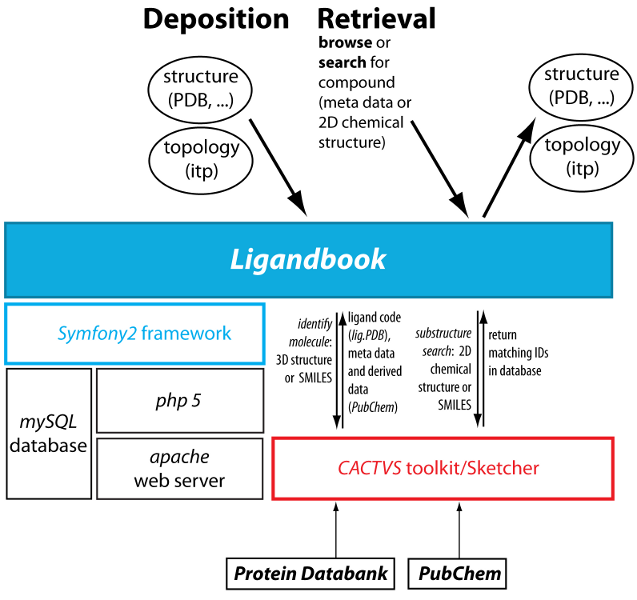
Ligandbook software architecture. Ligandbook is built on the PHP symfony2 framework and makes use of the cheminformatics CACTVS toolkit.
Ligandbook is written in PHP with the Symfony2 framework and uses MySQL as a relational database. Parameter files are stored as flat files on disk. The CACTVS toolkit has been interfaced with the PHP framework in order to provide cheminformatics services such as identifying compounds from 3D structures (as provided in input PDB files) or substructure search. The CACTVS Sketcher allows drawing of chemical structures as input for the structure search. Text search uses the elasticsearch engine that is built on top of the Apache Lucene Core search engine library.
Some of the quotations appearing on the site were taken from the Gromacs "cool quotes" (used under the GNU General Public Licence).
This work was supported through funding from Institut de Chimie des Substances Naturelles (ICSN), CNRS, Gif-sur-Yvette, France, Laboratory of Excellence in Research on Medication and Innovative Therapeutics (LERMIT), and Arizona State University, Tempe AZ, USA.